
Shulin Cao
"In the pursuit of light, we often find ourselves in the most beautiful shadows."
Swipe Up or Scroll to Explore
2025 / Present
Big Sur, California
Swipe Left to See Full Gallery →
Featured Research
Regulatory Network Interactions
2023 · American Journal of Physiology-Heart
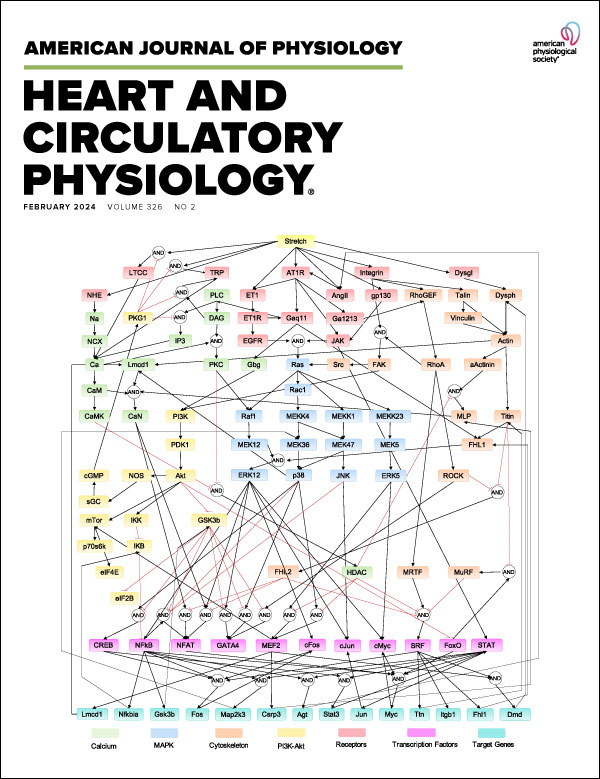
A Regulatory Network Model in PAH
2021 · Cells
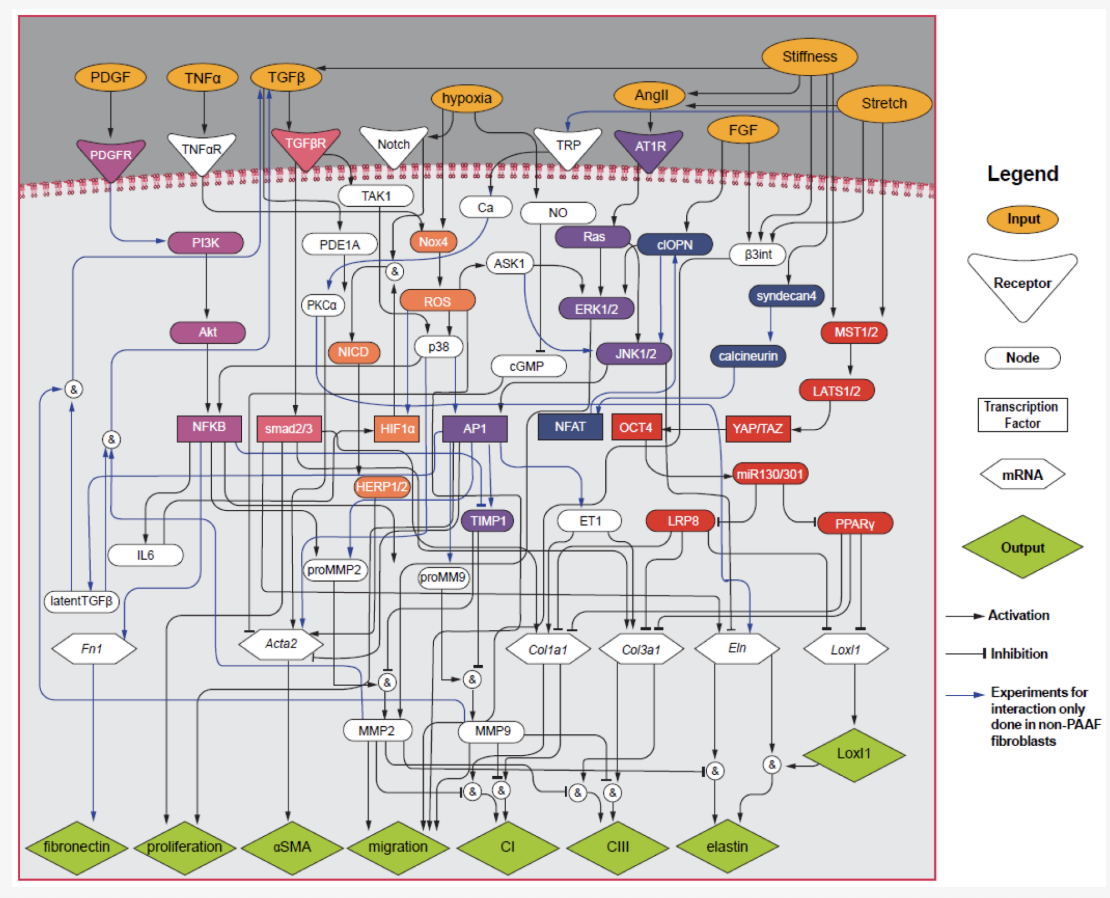
Interactivity in Network Regulation
2020 · Phil Trans R Soc A
Swipe Left for Full Archive →
Paper2023
Regulatory Network Interactions - A Biomedical Representation
Differential sensitivity to longitudinal and transverse stretch mediates transcriptional responses in mouse neonatal ventricular myocytes

Paper2021
A Regulatory Network Model in PAH
Substrate stiffness and stretch regulate profibrotic mechanosignaling in pulmonary arterial adventitial fibroblasts

Paper2020
Uncertainty Quantification in PAH
Quantification of uncertainty in a new network model of pulmonary arterial adventitial fibroblast pro-fibrotic signalling
Paper2020
Interactivity in Network Regulation
Quantification of model and data uncertainty in a network analysis of cardiac myocyte mechanosignalling

Big Sur, California


